Research
To investigate the molecular basis of biological photoreception we employ a combination of various techniques from molecular biology to advanced spectroscopy.
 Photoreceptors using flavins as pigments are fascinating objects of
study for several reasons. In contrast to other photoreceptor proteins
using isomerisable cofactors like carotenoids or tetrapyrrols they
undergo rather little structural transitions upon illumination.
Instead, they use classical flavoprotein chemistry to translate light
into biological information. Because of their reversible
photoswitching behavior they can be studied by reaction induced
spectroscopy with time resolution down to femtoseconds, which makes
them perfect model systems for flavoproteins in general.
Photoreceptors using flavins as pigments are fascinating objects of
study for several reasons. In contrast to other photoreceptor proteins
using isomerisable cofactors like carotenoids or tetrapyrrols they
undergo rather little structural transitions upon illumination.
Instead, they use classical flavoprotein chemistry to translate light
into biological information. Because of their reversible
photoswitching behavior they can be studied by reaction induced
spectroscopy with time resolution down to femtoseconds, which makes
them perfect model systems for flavoproteins in general.The blue light sensors using FAD (BLUF) represent a modular photoreceptor family that features the photosensing BLUF domain regulating the activity various enzymatic output domains. Especially the photoactivated adenylyl cyclases (PAC) have proven to be potent optogenetic tools to control cAMP induced signal transduction. The BLUF domain itself undergoes only a subtle hydrogen bond switch between the flavin and the protein upon illumination and the signal transduction to the cyclase domain in PACs, for example, is still poorly understood.
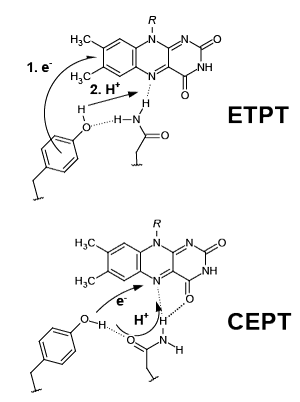
Furthermore, BLUF domains are powerful model systems to study proton coupled electron transfer (PCET). Upon illumination ultrafast electron transfer (ET) followed by proton transfer (PT) takes place from a conserved tyrosine to the flavin. The resulting neutral flavin semiquinone/tyrosine radical pair recombines within less than a nanosecond to facilitate the hydrogen bond switch. The nature of the photoinduced PCET is delicately modulated by the protein network.
Protein preparation
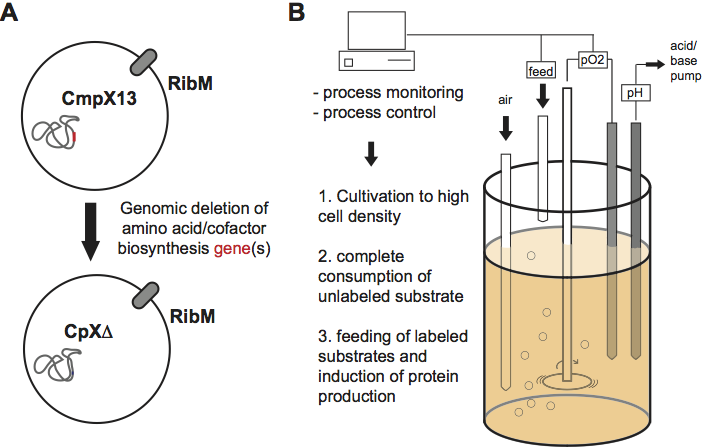 In order to investigate protein function with molecular resolution
proteins have to be available in high amounts and high purity. We use
Escherichia coli to
overproduce the proteins of interest with state-of-the-art expression
systems and purify them using affinity tags. However, for some
spectroscopic applications like vibrational or NMR spectroscopy these
proteins need to be selectively isotope labeled. For this purpose we
genomically engineered expression strains deficient in the
biosynthesis of selected amino acids or flavin cofactors (Mehlhorn et
al, 2013 PLOS ONE).
In order to investigate protein function with molecular resolution
proteins have to be available in high amounts and high purity. We use
Escherichia coli to
overproduce the proteins of interest with state-of-the-art expression
systems and purify them using affinity tags. However, for some
spectroscopic applications like vibrational or NMR spectroscopy these
proteins need to be selectively isotope labeled. For this purpose we
genomically engineered expression strains deficient in the
biosynthesis of selected amino acids or flavin cofactors (Mehlhorn et
al, 2013 PLOS ONE).  To meet the high demand for selectively labeled protein for
spectroscopy we use high cell density fermentation. The cultivation is
designed in way that unlabeled substrates are used exclusively for
biomass generation and labeled substrates only for the expression of
the protein of interest.
To meet the high demand for selectively labeled protein for
spectroscopy we use high cell density fermentation. The cultivation is
designed in way that unlabeled substrates are used exclusively for
biomass generation and labeled substrates only for the expression of
the protein of interest. 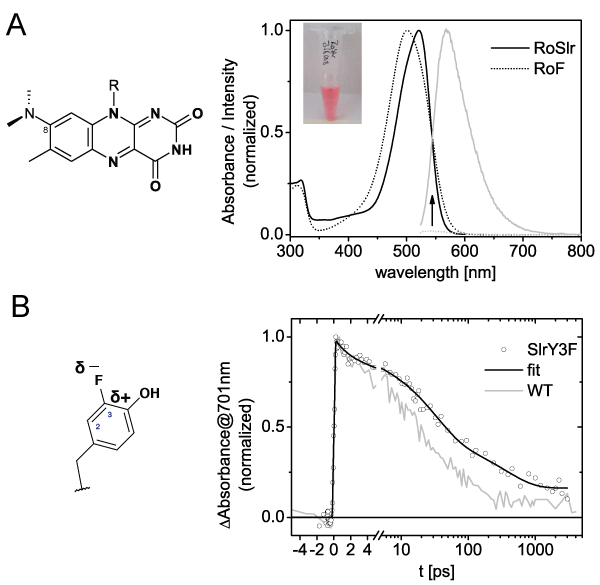
With this strategy we are not only able to introduce selective isotope labels into (flavo-)proteins but also to incorporate non-natural cofactors or amino acids (Kennis & Mathes, 2013 Interface FOCUS).
November, 2014

