|
|
|
|
|
|
2018 (54)
|
 |
Meike H. van der Ree, Louis Jansen, Matthijs R.A. Welkers, Hendrik W. Reesin, K. Anton Feenstra, and Neeltje A. Kootstra.
Deep sequencing identifies hepatitis B virus core protein signatures in chronic hepatitis B patients,
Antiviral Research in press (2018).
|  |
|
 |
| |
 |
Annika Jacobsen, Linda J. W. Bosch, Sanne R. Martens-De Kemp, Beatriz Carvalho, Anke H. Sillars-Hardebol, Richard J. Dobson, Emanuele De Rinaldis, Gerrit A. Meijer, Sanne Abeln, Jaap Heringa, Remond J.A. Fijneman, and K. Anton Feenstra.
Aurora kinase A (AURKA) interaction with Wnt and Ras-MAPK signalling pathways in colorectal cancer,
Scientific Reports 8 p. 7522 (2018).
|  |
|
 |
| |
 |
K. Anton Feenstra, Sanne Abeln, Johan A. Westerhuis, Filippe B. dos Santos, Douwe Molenaar, Bas Teusink, Huub J. C. Hoefsloot, and Jaap Heringa.
Training for translation between disciplines: a philosophy for life and data sciences curricula,
Bioinformatics 34 p. 4-12 (2018).
|  |
|
 |
| |
|
|
2017 (2)
|
 |
Punto Bawono, Maurits Dijkstra, Walter Pirovano, Anton Feenstra, Sanne Abeln, Jaap Heringa.
Multiple sequence alignment,
Methods in Molecular BIology 1525 p. 167-189 (2017).
|  |
 |
|
| |
 |
Qingzhen Hou, Paul de Geest, Wim F. Vranken, Jaap Heringa and K. Anton Feenstra.
Seeing the trees through the forest: sequence-based homo- and heteromeric protein-protein interaction sites prediction using random forest,
Bioinformatics 33 p. 1479-1487 (2017).
|  |
|
 |
| |
|
|
2016 (3)
|
 |
Qingzhen Hou, Marc F. Lensink, Jaap Heringa and K. Anton Feenstra.
CLUB-MARTINI: Selecting Favourable Interactions amongst Available Candidates, a Coarse-Grained Simulation approach to Scoring Docking Decoys,
PLoS ONE (2016) doi:10.1371/journal.pone.0155251.
|  |
|
 |
| |
 |
Annika Jacobsen, Nika Heijmans, Folkert Verkaar, Martine J. Smit, Jaap Heringa*, Renée van Amerongen* and K. Anton Feenstra*.
Construction and Experimental Validation of a Petri Net Model of Wnt/β-catenin Signaling,
PLoS ONE (2016) doi:10.1371/journal.pone.0155743.
* shared senior authors
|  |
|
 |
| |
 |
Reza Haydarlou, Annika Jacobsen, Nicola Bonzanni, K. Anton Feenstra, Sanne Abeln, Jaap Heringa.
BioASF: A Framework for Automatically Generating Executable Pathway
Models Specified in BioPAX,
Bioinformatics (2016) 32:i60-i69. doi: 10.1093/bioinformatics/btw250.
|  |
|
 |
| |
|
|
2015 (2)
|
 |
Qingzhen Hou, Bas E. Dutilh, Martijn A. Huynen, Jaap Heringa and K. Anton Feenstra.
Sequence specificity between interacting and non-interacting homologs identifies interface residues — a homodimer and monomer use case,
BMC Bioinformatics 16:325, (2015).
|  |
|
 |
| |
 |
Bas Stringer, Maurits Dijkstra, Anton Feenstra, Sanne Abeln, Jaap Heringa.
Explaining disease using big data: How valid is your pathway?,
in High Performance Computing & Simulation (HPCS), 2015 International Conference on pp.662-664, doi:10.1109/HPCSim.2015.7237114 (2015).
| |
 |
|
|
|
2014 (2)
|
 |
Nicola Bonzanni, K. Anton Feenstra,
Wan Fokkink and Jaap Heringa.
Petri Nets are a Biologist's Best Friend,
in: Proc. First International Symposium on Formal Methods in Macro-Biology - FMMB 2014.
Lecture Notes in Computer Science
(2014).
|  |
 |
 |
| |
 |
Ali May*, Rene Pool*, Erik van Dijk, Jochem Bijlard, Sanne Abeln, Jaap Heringa and K. Anton Feenstra
Coarse-grained vs. atomistic simulations: realistic interaction free energies for real proteins,
Bioinformatics (2014) 30: 326-334..
* shared first authors
|  |
 |
 |
|
|
2013 (6)
|
 |
Timo Willemsen, Paul T. Groth and K. Anton Feenstra
Building Exceutable Biological Pathway Models Automatically from BioPAX,
In 3rd International Workshop on Linked Science 2013—Supporting Reproducibility, Scientific Investigations and Experiments (LISC2013). Sydney. (2013).
|  |
 |
 |
| |
 |
Esther F. Gijsbers, K. Anton Feenstra, Ad C. van Nuenen,
Marjon Navis, Jaap Heringa, Hanneke Schuitemaker and Neeltje A. Kootstra
HIV-1 replication fitness of HLA-B*57/5801 CTL escape variants
is restored by the accumulation of compensatory mutations in Gag,
PLoS ONE 8:12 (2013) doi:10.1371/journal.pone.0081235.
|  |
 |
 |
| |
 |
Tom L.G.M. van den Kerkhof, K. Anton Feenstra, Zelda Euler, Marit J. van Gils, Linda W.E. Rijsdijk, Brigitte D. Boeser-Nunnink, Jaap Heringa, Hanneke Schuitemaker and Rogier W. Sanders
HIV-1 envelope glycoprotein signatures that correlate with the development of cross-reactive neutralizing activity,
Retrovirology 10:102 (2013) doi:10.1186/1742-4690-10-102.

|  |
 |
 |
| |
 |
Sanne Abeln, Douwe Molenaar, K. Anton Feenstra, Huub C. J. Hoefsloot, Bas Teusink and Jaap Heringa
Bioinformatics and Systems Biology: bridging the gap between heterogeneous student backgrounds,
Briefings in Bioinformatics, doi:10.1093/bib/bbt023 (2013).
|  |
|
 |
| |
 |
Nicola Bonzanni*, Abhishek Garg*, K. Anton Feenstra*, Sarah Kinston, Diego Miranda-Saavedra, Judith Schutte, Jaap Heringa, Ioannis Xenarios and Berthold Göttgens
Hard-wired heterogeneity in blood stem cells revealed
using a dynamic regulatory network model,
Bioinformatics, 29(13): i80-i88 (2013).
Best paper in Translational Bioinformatics at ISMB/ECCB 2013 award
* shared first authors
|  |
 |
 |
| |
 |
Ingrid J. De Vries-van Leeuwen, Daniel da Costa Pereira, Koen D Flach, Sander R Piersma, Christian Haase, David Bier, Zeliha Yalcin, Rob Michalides, K. Anton Feenstra, Connie R Jiménez, Tom F.A. de Greef, Luc Brunsveld, Christian Ottmann, Wilbert Zwart, Albertus H. de Boer
14-3-3 protein interaction with the Estrogen Receptor Alpha F-domain provides a drug target interface,
PNAS, 110(22): 8894-8899 (2013).
|  |
|
 |
|
|
2012 (1)
|
 |
René Pool, Jaap Heringa, Hoefling, M., Schulz, R., Smith, J.C., K. Anton Feenstra
Enabling grand-canonical Monte Carlo: Extending the flexibility of GROMACS through the GromPy python interface module,
J. Comput. Chem., 33(12): 1207-1214 (2012).
|  |
 |
 |
| |
2010 (3)
|
 |
Walter Pirovano, Sanne Abeln, K. Anton Feenstra, and Jaap Heringa
Multiple alignment of membrane protein sequences,
in
Structural Bioinformatics of Membrane Proteins, (D. Frishman, Ed.),
Springer: Wien, New York. ISBN: 9783709100448.
|  |
|
 |
| |
 |
Bernd W. Brandt*, K. Anton Feenstra*, and Jaap Heringa
Multi-Harmony: detecting functional specificity from sequence alignment,
Nucl. Acids Res., doi:10.1093/nar/gkq415 (2010).
* shared first authors
|  |
 |
 |
| |
 |
Lucas J. Gursky#*, Jasmina Nikodinovic-Runic#*,
K. Anton Feenstra and Kevin E. O'Connor#
In vitro evolution of styrene monooxygenase from Pseudomonas
putida CA-3 with a view to generating a better
biocatalyst,
Appl.
Microbiol. & Biotech., 85:p995-1004 (2010).
#School of Biomolecular and Biomedical Sciences,
Ardmore House, and the Centre for Synthesis and Chemical
Biology, University College Dublin, Ireland.
* shared first authors
|
 |
 |
 |
| |
|
|
2009 (3)
|
 |
Nicola Bonzanni, K. Anton Feenstra,
Wan Fokkink and Elzbieta Krepska.
What can Formal Methods bring to Systems Biology?,
in: Proc.
16th Symposium on Formal Methods - FM'09. pp. 16-22.
Lecture Notes in Computer Science 5850
(2009).
|
 |
 |
|
| |
 |
Walter Pirovano, Anneke van der Reijden, K. Anton Feenstra and Jaap Heringa.
Structure and Function Analysis of Flexible Alignment Regions in Proteins.,
BMC Bioinformatics 19:P6, (2009).
|  |
 |
 |
| |
 |
Nicola Bonzanni, Elzbieta Krepska, K. Anton Feenstra,
Wan Fokkink, Thilo Kielmann, Henri Bal and Jaap Heringa.
Executing Multicellular Development:
Quantitative Predictive Modelling of C. elegans Vulva,
Bioinformatics, 25:2049-2056 (2009).
|  |
 |
 |
| |
|
|
2008 (7)
|
 |
Walter Pirovano, K. Anton Feenstra and Jaap Heringa.
The meaning of alignment: lessons from structural diversity.,
BMC Bioinformatics, 9:556 (2008). 
|  |
 |
 |
| |
 |
K. Anton Feenstra, Giacomo Bastianelli# and Jaap Heringa.
Predicting Protein Interactions from Functional Specificity.
in: From
Computational Biophysics to Systems Biology. pp. 89-92.
Eds. U.H.E. Hansmann, J. Meinke, S. Mohanty and O. Zimmermann,
John von Neumann Institute for Computing, Jülich,
NIC Series, Vol. 40, 2008.
#Institut Pasteur, Paris, France. |
 |
 |
 |
| |
 |
Elzbieta Krepska, Nicola Bonzanni, K. Anton Feenstra,
Wan Fokkink, Thilo Kielmann, Henri Bal and Jaap Heringa.
Design Issues for Qualitative Modelling of Biological Cells
with Petri Nets,
Formal
Meth. in Syst. Biol., 5054:48-62 (2008).
|  |
 |
 |
| |
 |
Walter Pirovano, K. Anton Feenstra and Jaap Heringa.
PRALINETM: a strategy for improved multiple
alignment of transmembrane proteins,
Bioinformatics, 24:492-497 (2008).
|  |
 |
 |
| |
 |
Philip Lijnzaad#, K. Anton Feenstra, Jaap Heringa,
Frank C.P. Holstege#
On Defining the Dynamics of Hydrophobic Patches on Protein Surfaces,
Proteins,
72:105-114 (2008).
#Department of Physiological Chemistry, University Medical Center Utrecht |
 |
 |
 |
| |
 |
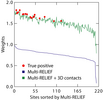
Kai Ye#, K. Anton Feenstra, Jaap Heringa,
Adriaan P. IJzerman#, Elena Marchiori
Multi-RELIEF: a method to recognize specificity determining
residues from multiple sequence alignments using a Machine
Learning approach for feature weighting,
Bioinformatics, 24: 18 - 25 (2008)
#Div. Medical Chemistry, LACDR, Leiden Univ., |
 |
 |
 |
| |
 |
K. Anton Feenstra, Chris de Graaf and Nico P. E.
Vermeulen Modeling of the active sites of Cyt P450s
in: Drug-drug interactions: second edition., Ed.
A. David Rodrigues, Informa Healthcare USA, Inc. (2008).
|  |
|
 |
| |
|
|
2007 (2)
|
 |
K. Anton Feenstra, Walter Pirovano, Klaas Krab# and Jaap Heringa
Sequence Harmony: Detecting Functional Specificity from Alignments,
Nucl. Acids Res., 35, W495-W498 (2007).
#Institute of Molecular Cell Biology, Vrije Universiteit Amsterdam |
 |
 |
 |
| |
 |
K. Anton Feenstra, Eugene B. Starikov, Vlada B. Urlacher#,
Jan N.M. Commandeur and Nico P.E. Vermeulen Combining Substrate
Dynamics, Binding Statistics and
Reactivity to Rationalize Regioselective Metabolism of Octane and
Lauric Acid by CYP102A1 and Mutants, Prot.
Sci., 16: 420-431 (2007).
# Institute of Technical Biochemistry, University of Stuttgart |
 |
 |
 |
| |
|
|
2006 (6)
|
 |
Elena Marchiori*, Walter Pirovano, Jaap Heringa and K. Anton Feenstra*.
A Feature Selection Algorithm for Detecting Subtype Specific Sites for Smad Receptor Binding,
Bio-ICMLA06, 168-173 (2006)
(IEEE).
* equal contribution |
 |
 |
 |
| |
 |
Walter Pirovano*, K. Anton Feenstra* and Jaap Heringa.
Sequence Comparison by Sequence Harmony Identifies Subtype
Specific Sites,
Nucleic Acids Res., 34, 6540-6548 (2006).
* shared first authors
|  |
 |
 |
| |
 |
K. Anton Feenstra, Karin Hofstetter#, Rolien Bosch,
Andreas Schmid#,¶, Jan N.M. Commandeur
and Nico P.E. Vermeulen
Enantioselective Substrate Binding in a Monooxygenase Protein
Model by Molecular Dynamics and Docking,
Biophys. J. 91 3206-3216 (2006)
# Chair of Chemical Biotechnology, Dept. of Biochemical and
Chemical Engineering, University of Dortmund, Germany
¶ ISAS-Institute for Analytical Sciences, Dortmund.
|
 |
 |
 |
| |
 |
K. Anton Feenstra, Walter Pirovano and Jaap Heringa.
Sub-type Specific Sites for SMAD Receptor Binding Identified
by Sequence Comparison using "Sequence Harmony".
in: From Computational Biophysics to Systems Biology.
pp. 73-78. Eds. U.H.E. Hansmann, J. Meinke, S. Mohanty and O. Zimmermann,
John von Neumann Institute for Computing, Jülich,
NIC Series, Vol. 34, 2006. |
 |
 |
 |
| |
 |
Oliver Lentz#, Anton Feenstra,
Tilo Habicher¶, Bernhard Hauer¶,
Rolf D. Schmid#, Vlada B. Urlacher#
Altering the Regioselectivity of Cytochrome P450 CYP102A3 of
Bacillus subtilis by Using a New Versatile Assay System,
ChemBioChem 7 345-350 (2006).
# Institut für Technische Biochemie, Universität
Stuttgart, Germany
¶ BASF AG, Ludwigshafen, Germany |
 |
 |
 |
| |
 |
Karin Hofstetter#,¶, Anton Feenstra,
Simon Alioth#, Jennifer Venhorst, Nico Vermeulen,
Bernard Witholt¶, Andreas Schmid#
Insight the catalytic site of styrene monooxygenase.
in: Biocatalytic asymmetric epoxidation of styrene and
derivatives using isolated styrene monooxygenase.
K. Hofstetter,
PhD Thesis nr. 16400, ETH, Zürich (2006).
# Dept. of Biochemical and Chemical Engineering,
University of Dortmund, Germany
¶ Institute of Biotechnology, HPT, Swiss Federal
Institute of Technology Zürich (ETHZ) |
 |
 |
 |
|
|
2005 (3)
|
 |
Peter H.J. Keizers, Chris de Graaf, Frans J.J.de Kanter,
Chris
Oostenbrink, K. Anton Feenstra, Nico P.E. Vermeulen, and Jan N.M.
Commandeur Metabolic Regio- And Stereoselectivity Of Cytochrome
P450 2D6 Towards 3,4-methylenedioxy-N-alkyl-amphetamines: In Silico
Predictions And Experimental Validation J. Med. Chem.,
48 6117-27 (2005). |
 |
 |
 |
| |
 |
Chris de Graaf*, Nico P. E. Vermeulen and K. Anton Feenstra*
Cytochrome P450 in Silico: An Integrative Modeling Approach
J. Med. Chem.,
48 (8) 2725-55 (2005).
* equal contribution |
 |
 |
 |
| |
 |
Nico P.E. Vermeulen, Chris de Graaf,
Anton F. Feenstra, Chris Oostenbrink
Automated docking and virtual screening of substrates of cytochromes P450
(2005) Drug Metab. Rev. 37, 13-13 (28 Suppl. 1).
|
|
|
|
|
2004 (3)
|
 |
David van der Spoel, Erik Lindahl, Berk Hess, Aldert R. van
Buuren, Emile Apol, Pieter J. Meulenhoff, D. Peter Tieleman, A. L. T.
M. Sijbers, K. Anton Feenstra, Rudi van Drunen and Herman J. C.
Berendsen Gromacs User Manual version 3.2 (2004)
www.gromacs.org
|
 |
|
 |
| |
 |
Nico P.E. Vermeulen, Peter Keizers,
Marola M.H. van Lipzig, Chris de Graaf, Jennifer Venhorst,
Anton E. Feenstra, Jan N.M. Commandeur
In silico prediction and experimental validation of drug
binding and metabolism: The case of Cytochromes P450
(2004) Eur. J. Pharm. Sci. 23, S9-S9.
|
|
|
| |
 |
Nico P.E. Vermeulen, Chris de Graaf,
Marola M.H. van Lipzig MMH, Anton F. Feenstra
Automated docking and molecular dynamics simulations of
substrate binding in cytochromes P450
(2004) Drug Metab. Rev. 36, 15-15 (29 Suppl. 1).
|
|
|
|
|
2002 (3)
|
 |
K. Anton Feenstra, C. Peter#, Ruud M. Scheek,
Alan E. Mark and Wilfred F. van Gunsteren#
A comparison of methods for calculating NMR cross-relaxation
rates (NOESY and ROESY intensities) in small peptides. (2002)
J. Biomol. NMR 23, 181-194.
# Laboratory of Physical Chemistry, Swiss Federal Institute of
Technology Zürich (ETHZ) |
 |
 |
 |
| |
 |
Petra A. W. van den Berg#,*, K. Anton Feenstra*,
Arie van Hoek#, Alan E. Mark, Herman J. C. Berendsen and
Antonie J. W. G. Visser#
Dynamic conformations of flavin adenine dinucleotide:
simulated molecular dynamics of the flavin cofactor related to the
time-resolved fluorescence characteristics. (2002)
J. Phys. Chem. B 106, 8858-8869
# MicroSpectroscopy Centre, Laboratory of Biochemistry
Wageningen University
* shared first authors |
 |
 |
 |
| |
 |
K. Anton Feenstra
Long Term Dynamics of Proteins and Peptides.
PhD Thesis, Rijksuniversiteit Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands (2002)
|  |
|
 |
|
|
2001 (1)
|
 |
David van der Spoel, Aldert R. van Buuren, Emile Apol,
Pieter J. Meulenhoff, D. Peter Tieleman, A. L. T. M. Sijbers,
Berk Hess, K. Anton Feenstra, Erik Lindahl, Rudi van Drunen and
Herman J. C. Berendsen Gromacs User Manual version 3.0 (2001)
www.gromacs.org
|
 |
|
 |
|
|
1999 (1)
|
 |
K. Anton Feenstra, Berk Hess and Herman J. C.
Berendsen. Improving
Efficiency of Large Time-scale Molecular Dynamics
Simulations of Hydrogen-rich Systems. (1999) J. Comput. Chem.
20 (8), 786-798 |
 |
 |
 |
|
|
1998 (2)
|
 |
David van der Spoel, Aldert R. van Buuren, Emile Apol,
Pieter J. Meulenhoff, D. Peter Tieleman, Alfons L. T. M.
Sijbers, Berk Hess, K. Anton Feenstra, Rudi van Drunen and
Herman J. C. Berendsen
Gromacs User Manual version 2.0 Nijenborgh 4, 9747 AG
Groningen, The Netherlands (1998)
md.chem.rug.nl
|
 |
|
 |
| |
 |
K. Anton Feenstra and Herman J. C. Berendsen. The
domain
decomposition of a single-domain protein.
in: Monte Carlo approach to biopolymers and protein folding.
pp. 255-267. Eds. P. Grassberger, G.T. Barkema and W. Nadler,
Höchstleistungsrechenzentrum Jülich, Germany; World
Scientific, London, 1998. |
 |
 |
 |
|
|
1996 (1)
|
 |
David van der Spoel, K. Anton Feenstra, Marcus A.
Hemminga
and Herman
J. C. Berendsen. (1996). Molecular Modelling of the RNA Binding
N-terminal part of CCMV coat protein in Solution with Phosphate
Ions. Biophys. J. 71, 2920-2932. |
 |
 |
 |
| |









































































































